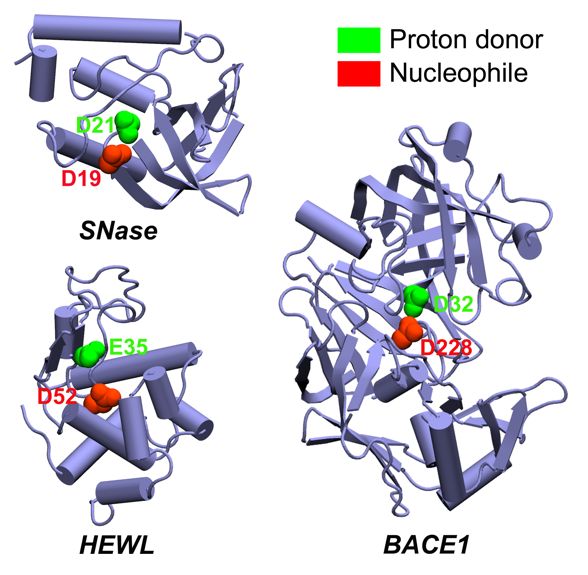
Constant pH Molecular dynamics
Extending thermodynamic ensembles to include constant solution pH has been a long-standing goal in the advancement of molecular dynamics (MD) methodologies. Continuous constant pH MD (CpHMD) makes use of the λ-dynamics based extended Hamiltonian to propagate a set of continuous titration coordinates.
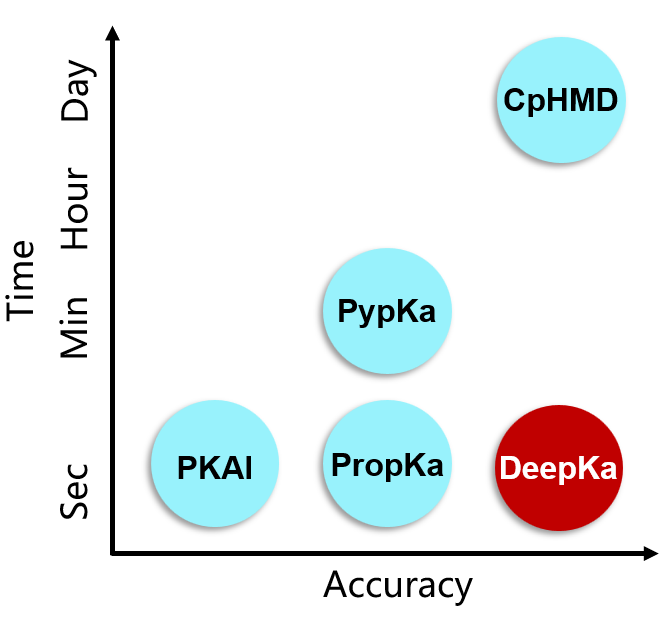
Protein pKa prediction
We developed the first working deep-learning based protein pKa predictor DeepKa, which is established by the data sets generated by continuous constant pH molecular dynamics simulations of 279 soluble proteins that include 12809 pKa values of four residue types, namely Asp, Glu, His and Lys. Notably, to avoid discontinuities at the boundary, grid charges are proposed to represent protein electrostatics. We show that the prediction accuracy by DeepKa is close to that by CpHMD benchmarking simulations, validating DeepKa as an efficient protein pKa predictor. In addition, the training and validation sets created in this study can be applied to the development of machine learning-based protein pKa predictors in the future. Finally, the grid charge representation is general and applicable to other topics, such as the protein-ligand binding affinity prediction. (Data and code can be downloaded for free from https://gitlab.com/yandonghuang/deepka.)
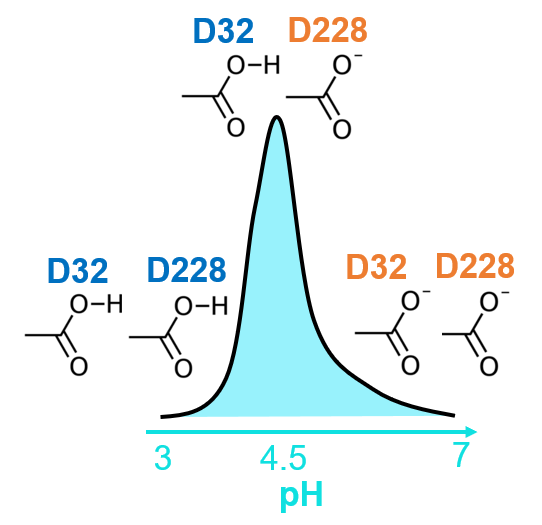
pH-dependent enzyme catalysis
Glycoside hydrolases (GHs) commonly use the retaining or inverting mechanisms to hydrolyze carbohydrates, and the rates of catalysis are usually pH dependent. Deeper understanding of these pH-dependent reaction mechanisms is of great importance for protein engineering and drug design.
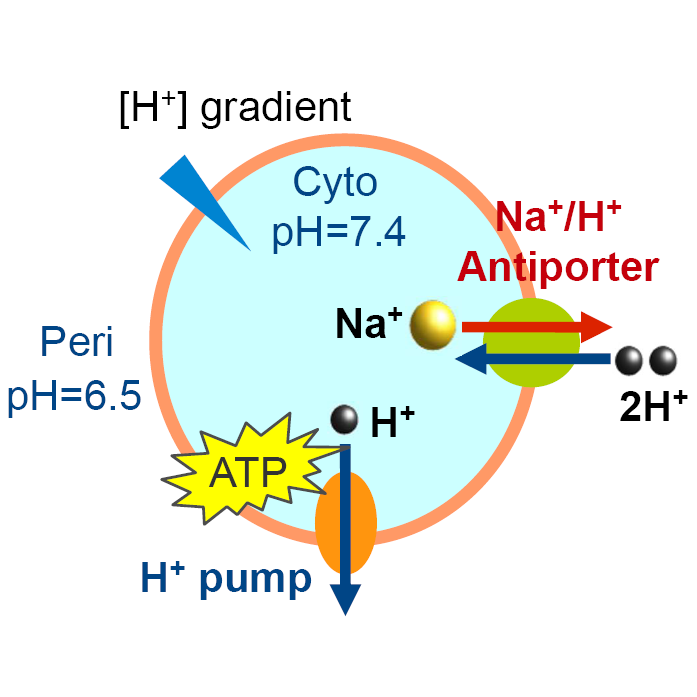
Proton-coupled matter transport
A lot of transmembrane proteins, such as sodium-proton antiporters, voltage-gated proton channels, and cholesterol transporters, function in coupled with solution pH. Thus it's of importance to explore pH-mediated mechanisms of these proteins.
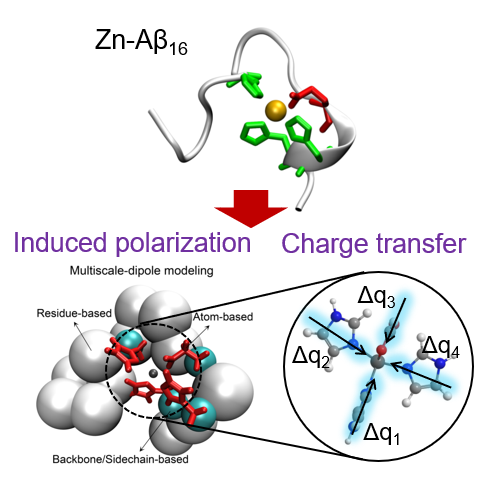
Zn protein simulation
Zinc can affect the structures and functions of many biomolecules that are related to human diseases, such as zinc finger protein, amyloid-beta peptide and voltage-gated proton channel Hv1. In order to better elucidate the interaction between zinc and a protein during the molecular dynamics simulation, induced polarization and charge transfer induced by zinc have been added to the classic force field. In particular, the zn-induced dipoles are divided into three distinct scales according to the distances between residues and zinc.
Ion channel gating dynamics
The propagation of action potentials is considered the most important type of neuronal signaling. Generation of action potentials and their transmission through the axon are governed by currents through ion channels on the neuronal membrane. Hodgkin and Huxley first described the nerve membrane ion currents deterministically and established what is now called the Hodgkin–Huxley (HH) mode. Stochasticity in ion channel gating is the major source of intrinsic neuronal noise, which can induce many important effects in neuronal dynamics. Several numerical implementations of the Langevin approach have been proposed to approximate the Markovian dynamics of the Hodgkin-Huxley neuronal model.
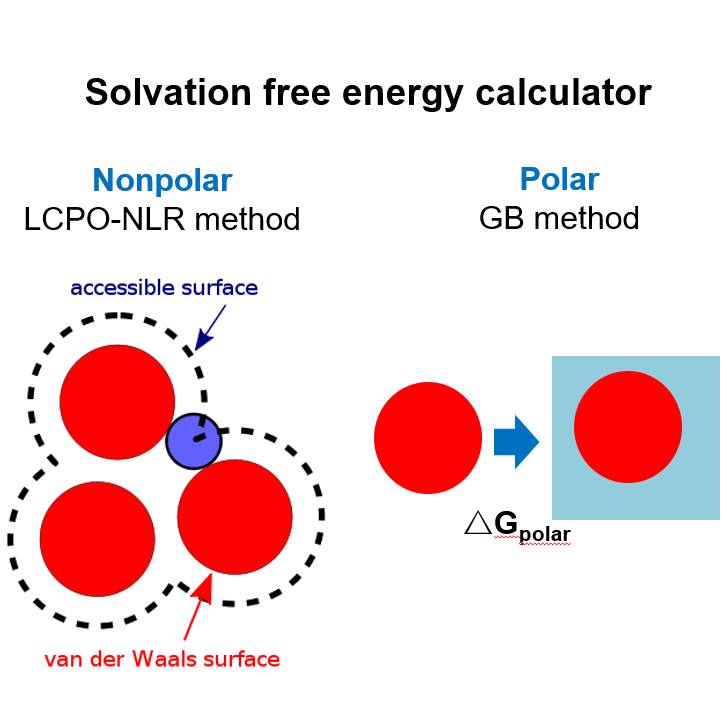
Solvation free energy calculation
We have set up an online service that provides the calculation of solvation free Energy of a protein. The solvation energy includes polar and nonpolar interactions. The polar part elucidates the electrostatic interaction between solute and solvent, which Is the main contributor to the solvation free energy and is estimated by Generalized-Born (GB) implicit solvent model in this service. The nonpolar indicates the hydrophobic interaction between solvent and solute atoms on the surface, which can be approximated by the calculation of the solvent accessible surface area (SASA) multiplied by a constant that represents the surface tensor. Users simply provide the protein pdb ID and the server will feedback with the corresponding SASA of the molecule.